- navigate_next Services
- navigate_next About SPILLO-PBSS
- navigate_next Experimental validations / Publications

We identify target and off-target proteins
of any small molecule on a proteome-wide scale
HIGH EFFICIENCY
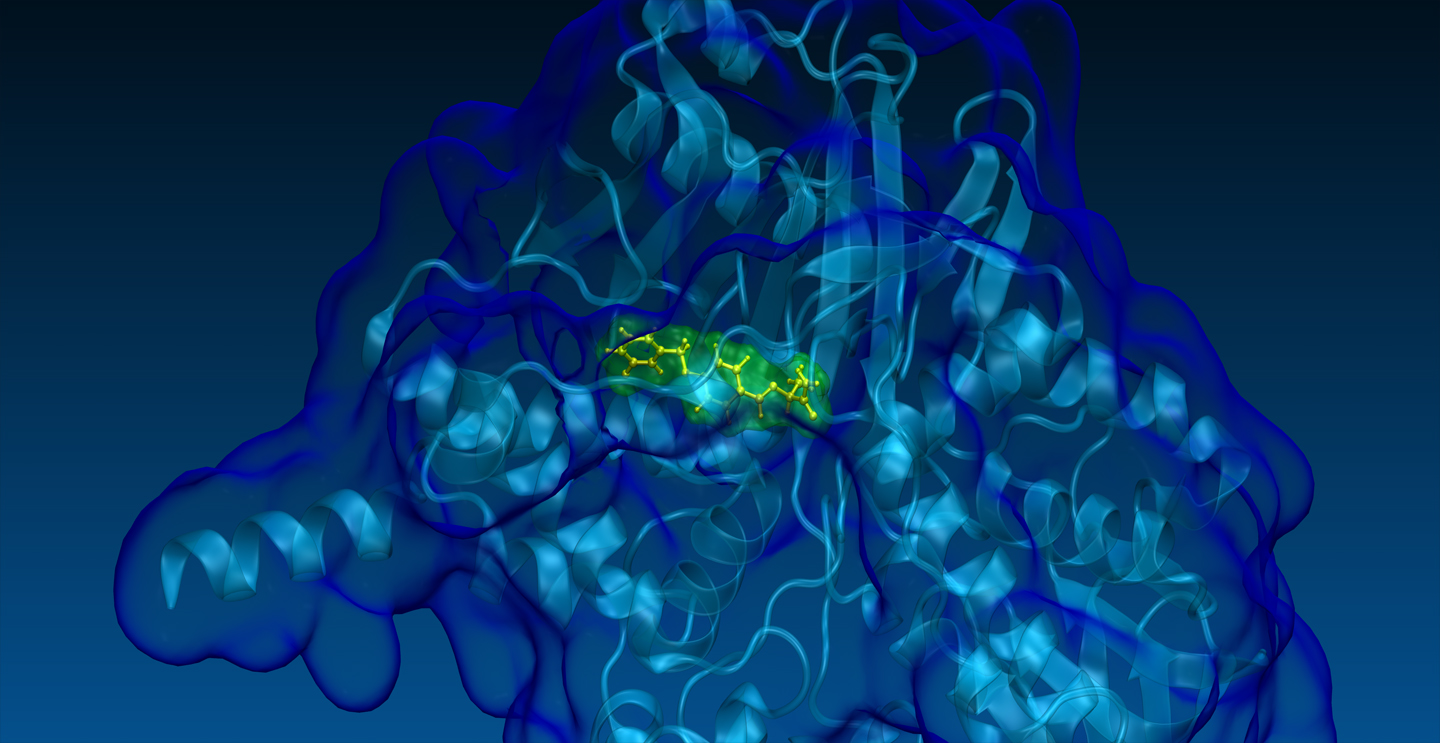
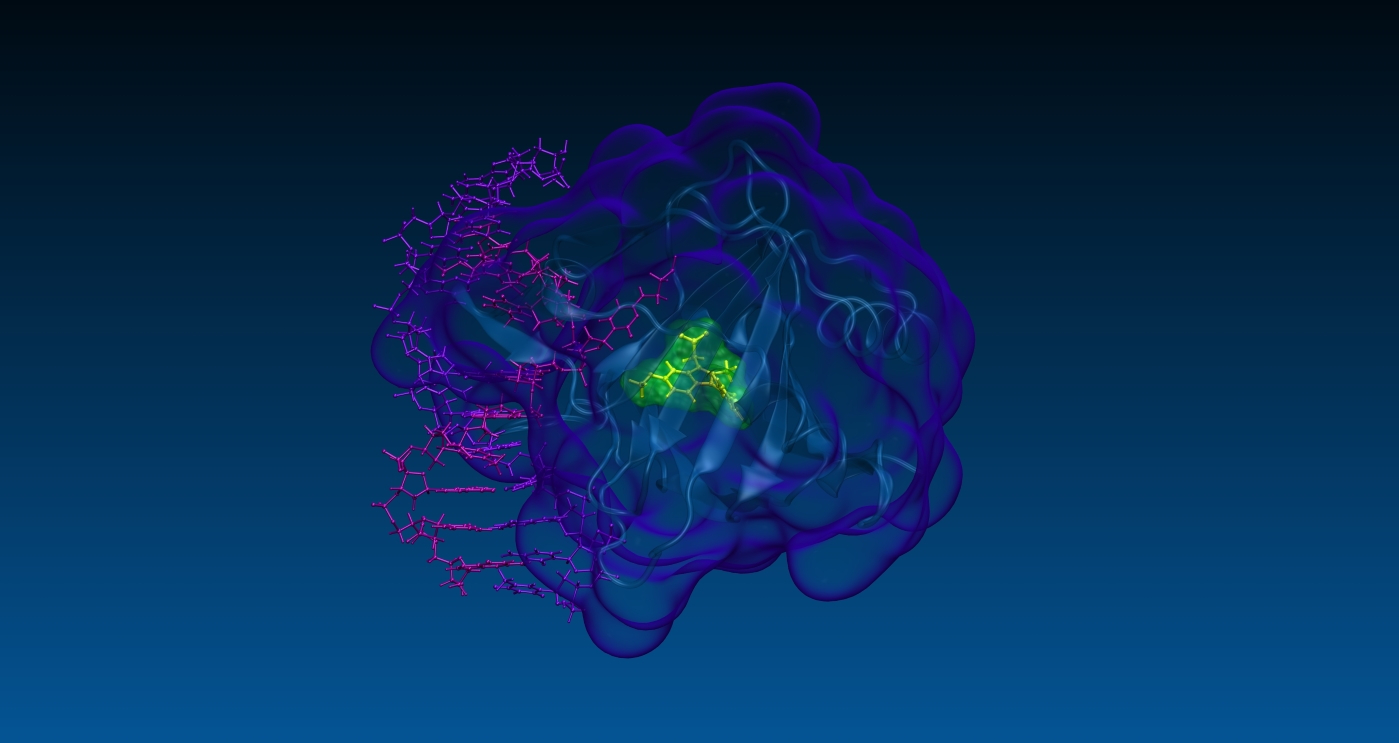
Our technologies allow to identify target and off-target proteins of any drug through a systematic analysis and ranking of very large protein databases, such as the whole available human structural proteome.
INNOVATION
&
UNIQUENESS
Our tools have unique potentialities in identifying drug binding sites on protein 3D-structures whether or not they are occupied, strongly distorted, or even completely closed, compared to a suitable conformation for the binding.
EXPERIMENTAL VALIDATIONS
To validate our approach while ensuring maximum objectivity and reliability, we have used our technology to tackle scientific problems still waiting to be solved. The results achieved have been experimentally confirmed by academic research centres and published in peer-reviewed scientific journals.
Developing and using new computational technologies for clinical pharmacology
flare
Clarifying ADVERSE EFFECTS
Adverse effects arising from aspecific drug-target interactions may be predicted and/or clarified at a biomolecular level through direct identification of the off-target proteins on a proteome-wide scale. The whole available human structural proteome can be screened by SPILLO-PBSS in affordable computation time.
filter_center_focus
Advancing PRECISION MEDICINE
Dose limitation and discontinuation of treatments due to adverse effects may expose patients to major health risks. A clearer drug-target interactions map can be generated by SPILLO-PBSS, aimed at designing safer and more effective therapies, according to individual patient’s variations.
grain
Promoting POLYPHARMACOLOGY
New drug-target interactions responsible for additional positive effects of a therapeutic agent may be successfully identified by SPILLO-PBSS. Polypharmacological therapies able to interfere with multiple targets of the same disease network may then be selected to improve the efficacy of the treatment.
lightbulb_outline
Clarifying THERAPEUTIC EFFECTS
If the mechanism of action of a therapeutic agent is not well-defined or still unknown, a screening of the whole available human structural proteome by SPILLO-PBSS can be crucial to identify the primary target responsible for the reported effects of that molecule.
redo
Facilitating DRUG REPOSITIONING
SPILLO-PBSS may find off-target proteins involved in pathological network other than that to which the primary target belongs, thus providing the opportunity for using known drugs for new indications, maximizing in this way the potential of a drug.
fullscreen_exit
Addressing RARE DISEASES
SPILLO-PBSS can be a valuable resource for neglected- and rare-diseases research in the effort to identify opportunities of drug repositioning, which represents a cheaper and faster alternative to traditional drug discovery.
Developing and using new computational technologies for drug R&D
A brief introduction to SPILLOproject
SPILLOproject is an Italian-based research and consulting firm, founded by Dr. Alessandro Di Domizio in 2016 with the goal of improving human health and advancing disease research, including rare and neglected diseases, by developing and using innovative computational technologies. The project is carried on by its founder together with an interdisciplinary team of experts.
Our in silico methodologies represent a unique and valuable resource (see peer-reviewed article in JCC) for all areas where molecular recognition via protein-ligand interactions plays a key role, including (among others) drug R&D, clinical pharmacology, precision medicine and toxicology.
We provide services to drastically reduce the cost of the drug discovery and development process, while increasing the probability of success of new therapies. We collaborate with private and public research centers, academia, pharmaceutical and biotech companies. We are at your disposal to answer any question concerning possible applications of our technologies to problems of your interest, and to carry out preliminary feasibility studies.
In evidence
-
bookmarkThree-Dimensional Proteome-Wide Scale Screening for the 5-Alpha Reductase Inhibitor Finasteride: Identification of a Novel Off-TargetJOURNAL OF MEDICINAL CHEMISTRY (2021) - Giatti S, Di Domizio A, Diviccaro S, Falvo E, Caruso D, Contini A, and Melcangi RC
Read full article -
bookmark3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell lineBIOORGANIC & MEDICINAL CHEMISTRY (2020) - Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V, and Nicolini G
Read full article -
bookmarkTubulin binding potentially clears up Bortezomib and Carfilzomib differential neurotoxic effectSCIENTIFIC REPORTS (2021) - Malacrida A, Semperboni S, Di Domizio A, Palmioli A, Broggi L, Airoldi C, Meregalli C, Cavaletti G, and Nicolini G
Read full article -
bookmarkMV1035 Overcomes Temozolomide Resistance in Patient-Derived Glioblastoma Stem Cell LinesBIOLOGY (2022) - Malacrida A, Di Domizio A, Bentivegna A, Cislaghi G, Messuti E, Tabano SM, Giussani C, Zuliani V, Rivara M, and Nicolini G
Read full article -
bookmarkIdentification of a novel off-target of paroxetine: possible role in sexual dysfunction induced by this SSRI antidepressant drug.JOURNAL OF MOLECULAR STRUCTURE (2022) - Giatti S, Di Domizio A, Diviccaro S, Cioffi L, Marmorini I, Falvo E, Caruso D, Contini A, and Melcangi RC
Read full article -
bookmarkFraisinib: a calixpyrrole derivative reducing A549 cells-derived NSCLC tumor in-vivo, acts as ligand of the Glycine-tRNA Synthase, a new molecular target in oncologyFRONTIERS IN PHARMACOLOGY (2023) - Ben Toumia I, Bachetti T, Chekig-Ghedira L, Profumo A, Ponassi M, Di Domizio A, Izzotti A, Sciacca S, et al.
Read full article
