- navigate_next Services
- navigate_next About SPILLO-PBSS
- navigate_next Experimental validations / Publications
Comparison with traditional structure-based approaches
SPILLO-PBSS has been compared with other available tools which could be used for the prediction of binding sites within protein 3D-structures, each tool being representative of a different class of methods.
Tests were performed on limit cases concerning the conformation of the binding site. In particular, a search for the binding site of the ligand 'Guanosine diphosphate (GDP)' was performed within four H-Ras protein conformations, in which various levels of distortion of the binding site are taken into account: (i) SUITABLY OPEN (undistorted); (ii) WIDE-OPEN; (iii) WIDE-OPEN and OCCUPIED; (iv) COMPLETELY CLOSED.
Results are reported in Table 2.
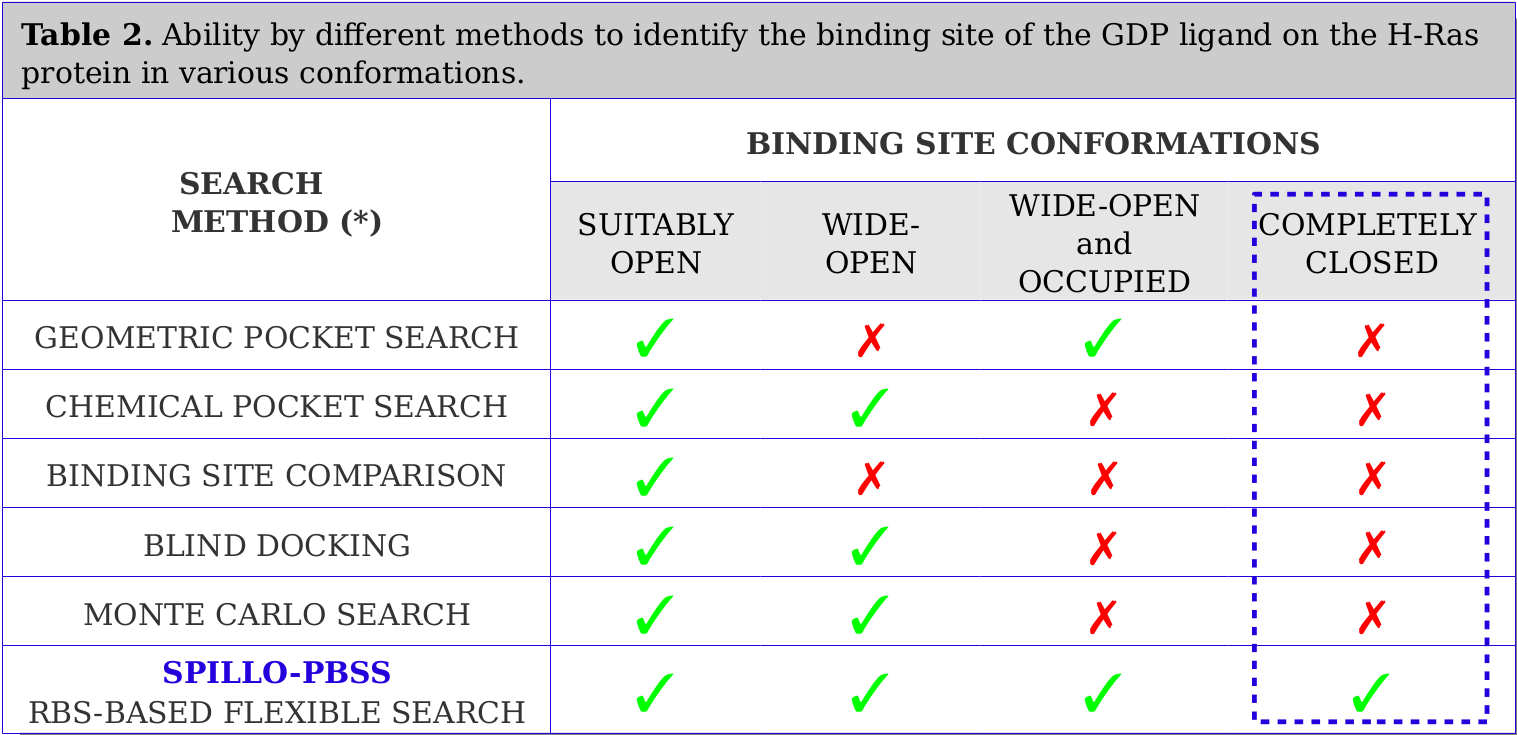
SPILLO-PBSS has proved successful in recognizing binding sites even within really distorted proteins. No method (apart from SPILLO-PBSS) has ever been able to recognize the expected binding site when COMPLETELY CLOSED and apparently INACCESSIBLE to the ligand (see the Video below).
SPILLO-PBSS ability in identifying the GDP (in yellow) binding site even when it is strongly distorted (i.e., fully closed). Both the surface and the inner regions of the RAS protein are analyzed, including those considered 'inaccessible' according to traditional methods.
*) For full details see the reference paper: SPILLO-PBSS: Detecting hidden binding sites within protein 3D-structures through a flexible structure-based approach, A. Di Domizio et al, Journal of Computational Chemistry, 2014, 35 (27): 2005-2017. DOI: 10.1002/jcc.23714
SPILLO-PBSS analyzes both empty (in blue) and occupied regions of the protein structure. It takes into account protein flexibility in solution and identifies binding sites even when the protein is not in a conformation suitable for the binding. This video shows a binding site where most of the small molecule is interpenetrated with the atoms of the protein. This result is strongly prohibited (apparently not realistic situation) by the other methods and would have been immediately discarded.
The target protein identified through the direct identification of this binding site has been experimentally validated (for more details, please see here).
